Archives
- 2025-11
- 2025-10
- 2025-09
- 2025-03
- 2025-02
- 2025-01
- 2024-12
- 2024-11
- 2024-10
- 2024-09
- 2024-08
- 2024-07
- 2024-06
- 2024-05
- 2024-04
- 2024-03
- 2024-02
- 2024-01
- 2023-12
- 2023-11
- 2023-10
- 2023-09
- 2023-08
- 2023-07
- 2023-06
- 2023-05
- 2023-04
- 2023-03
- 2023-02
- 2023-01
- 2022-12
- 2022-11
- 2022-10
- 2022-09
- 2022-08
- 2022-07
- 2022-06
- 2022-05
- 2022-04
- 2022-03
- 2022-02
- 2022-01
- 2021-12
- 2021-11
- 2021-10
- 2021-09
- 2021-08
- 2021-07
- 2021-06
- 2021-05
- 2021-04
- 2021-03
- 2021-02
- 2021-01
- 2020-12
- 2020-11
- 2020-10
- 2020-09
- 2020-08
- 2020-07
- 2020-06
- 2020-05
- 2020-04
- 2020-03
- 2020-02
- 2020-01
- 2019-12
- 2019-11
- 2019-10
- 2019-09
- 2019-08
- 2019-07
- 2019-06
- 2019-05
- 2019-04
- 2018-11
- 2018-10
- 2018-07
-
The electrocardiogram showed narrow QRS complex regular tach
2019-05-24
The electrocardiogram showed narrow QRS complex regular tachycardia, which was terminated by rapid injection of adenosine triphosphate. During the tachycardia, the P wave was not identifiable and was likely hidden completely in the QRS complex (Fig. 1). As the patient desired complete cure of the ta
-
3-Methyladenine br Adverse events Treatment reported toxicit
2019-05-24
Adverse events Treatment reported toxicities were similar to those previously reported [23]. Discussion Several other studies have addressed the same issue using different bisphosphonates. The SABRE study randomised breast cancer patients taking anastrozole with a T-score of between −1 and −2
-
Studies and reports of lung
2019-05-24
Studies and reports of lung fiin of GCTB are rare because of the low incidence of lung metastasis. The biological behavior and clinical features of GCTB are difficult to predict [6,7]. Some researchers have attempted to analyze related clinical factors of lung metastasis, such as age, sex, primary t
-
Understanding and defining these miRNA profiles
2019-05-24
Understanding and defining these miRNA profiles helps to uncover the clinical usefulness of miRNAs. Numerous human clinical trials are currently evaluating miRNA therapeutics during various liver diseases, such as targeting miR-122 during chronic hepatitis C infection. Aside from therapeutics, the d
-
Una recopilaci n impresa del trabajo que realiz con su
2019-05-24
Una recopilación impresa del trabajo que realizó con su cámara, desde 1980 hasta 1986, fue publicado con el título: Guatemala: Eternal Spring, Eternal Tyranny en 1988. En 2010 reeditó el libro en español, el cual se difundió ampliamente en Guatemala como parte del proceso de recuperación histórica.
-
Introduction By catalyzing post translational
2019-05-24
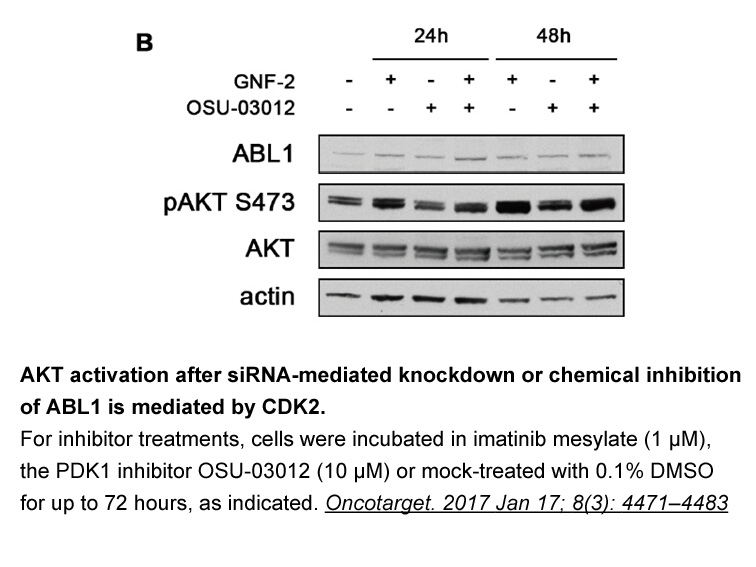
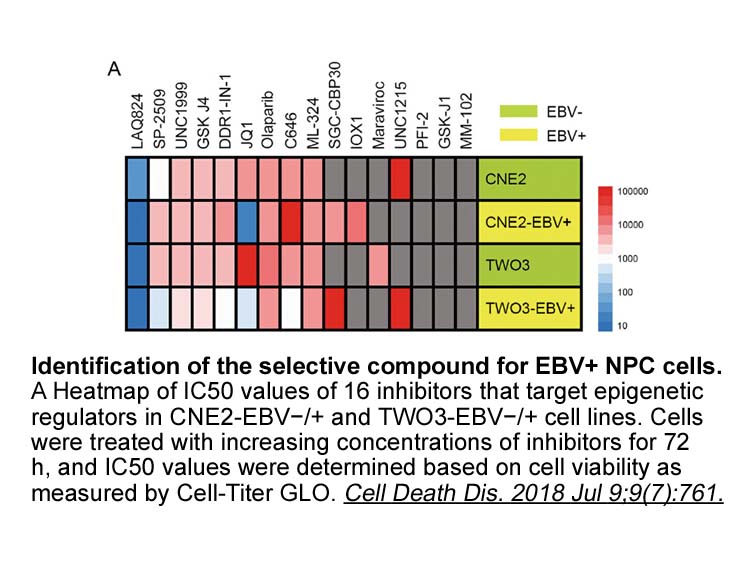
Introduction By catalyzing post-translational acetylation of lysine residues, the histone acetyl transferases (HAT) CREBBP and EP300 moderate the activities of both histone and non-histone proteins. This regulates vital cellular processes such as cell cycling and apoptosis by modulating chromatin s
-
Cancer stem cells are capable
2019-05-24
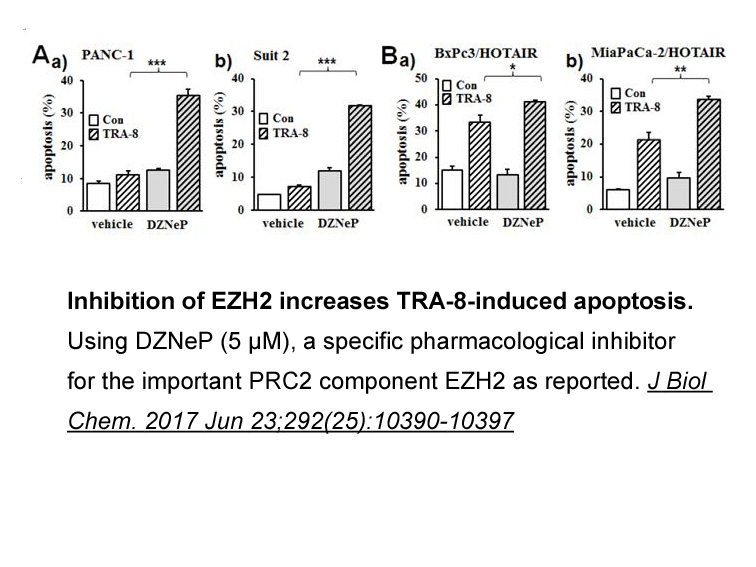
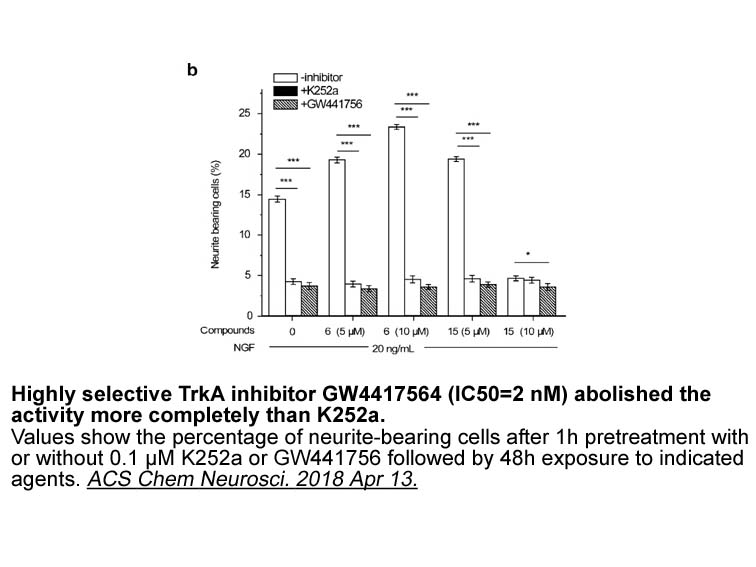
Cancer stem herpes simplex virus infection are capable of differentiation and self-renewal. In 1994, CSCs were described for the first time. Hematological malignance is the best evidence to support the existence CSCs. TPC (tumor Propagating cell) is the other phrase that has been used for cancer ste
-
Las pol ticas cosmopolitas de Or genes
2019-05-23
Las políticas cosmopolitas de Orígenes, por su parte, combinaron los intereses de sus directores, José Lezama Lima y José Rodríguez Feo. El primero, estaba empeñado en la formación de un grupo poético creativo, que trabajara en el diseño de una teleología para el proyecto republicano. Esta teología
-
br Conflict of interest br
2019-05-23
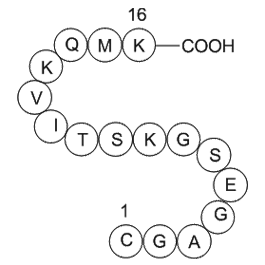
Conflict of interest Introduction Baseline ablation strategies targeting PVs It is well known that PVI was first developed to eliminate the triggers that initiate attacks of paroxysmal atrial fibrillation [1]. Subsequently, the additional function of the PV myocardium to perpetuate atrial f
-
Un a o despu s como parte del influjo posconciliar
2019-05-23
Un año después, como parte del influjo posconciliar, fue publicada la encíclica del papa Pablo vi, quien presidió su papado de 1963-1978, Populorum Progressio (1967). La novedad de este documento de enseñanza social católica fue la insinuación de una fuerte crítica hacia el orden económico internaci
-
br El fuego como fuerza destructora end gena
2019-05-23
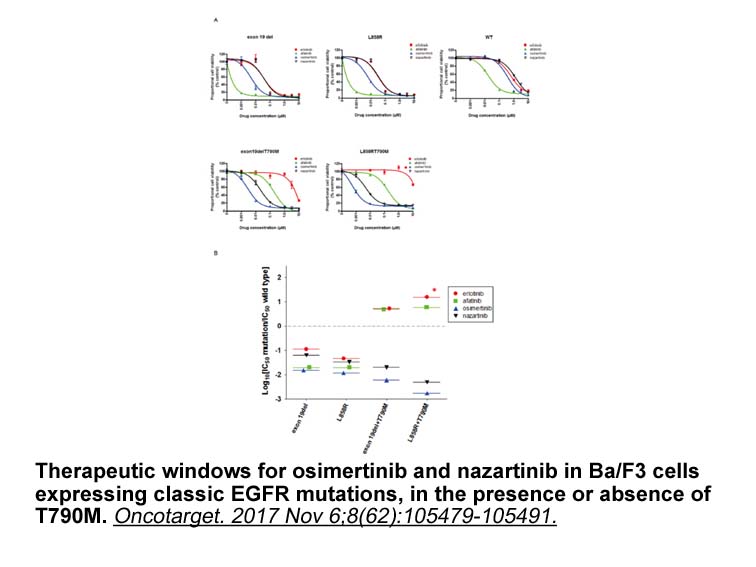
El fuego como fuerza destructora endógena Según Elizabeth Monasterios Pérez, la poesía de Pacheco, al igual que la de Octavio Paz, por ejemplo, se nutre de cosmología prehispánica. Con todo, prosigue Monasterios Pérez, Pacheco se demarca de Paz mediante la configuración de un “principio apocalípt
-
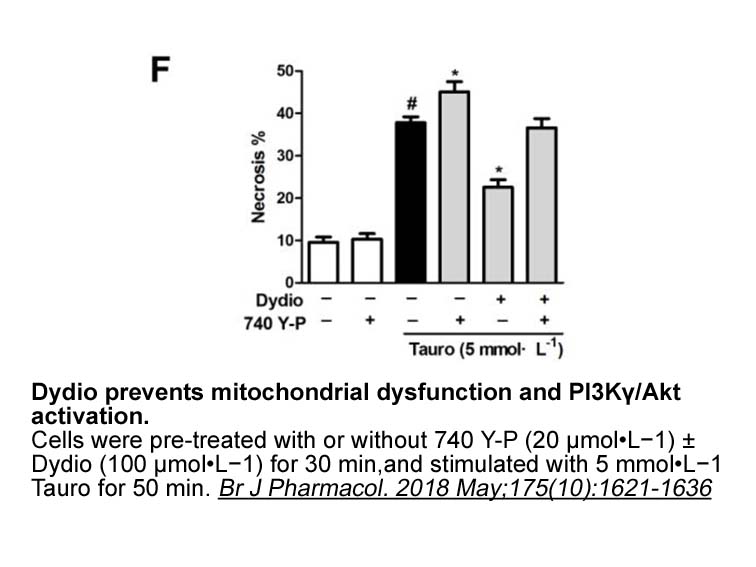
Another problem is that the indications of each drug
2019-05-23
Another problem is that the indications of each drug differ among Asia-Pacific countries, and it dcb is difficult to provide recommendations that are consistent with the indications in each country. For example, in Japan, the 3 novel OACs are indicated for “the prevention of ischemic stroke and sys
-
br Discussion The presence of
2019-05-23
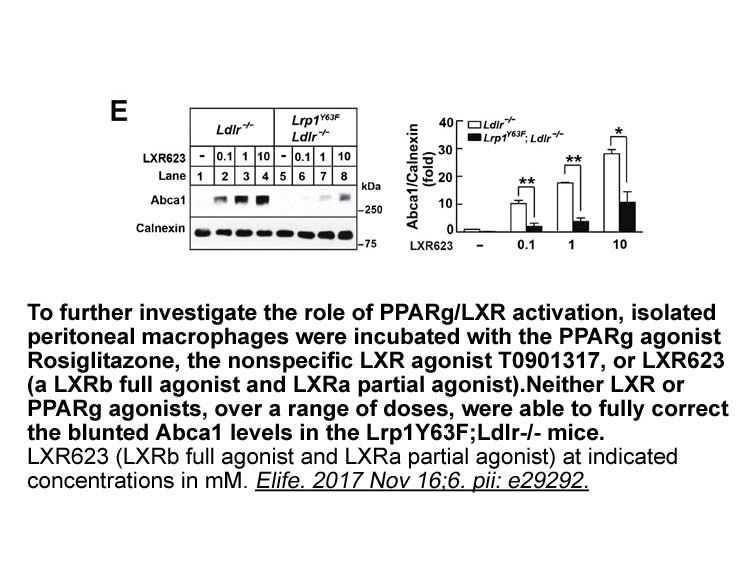
Discussion The presence of metastases at the time of diagnosis is a significant prognostic factor in osteosarcoma [14]. Skeletal metastases at time of presentations appears to carry a worse prognosis than pulmonary metastases [15]. Furthermore, the chance of survival can be improved by surgical r
-
br The Global Burden of Disease Study
2019-05-23
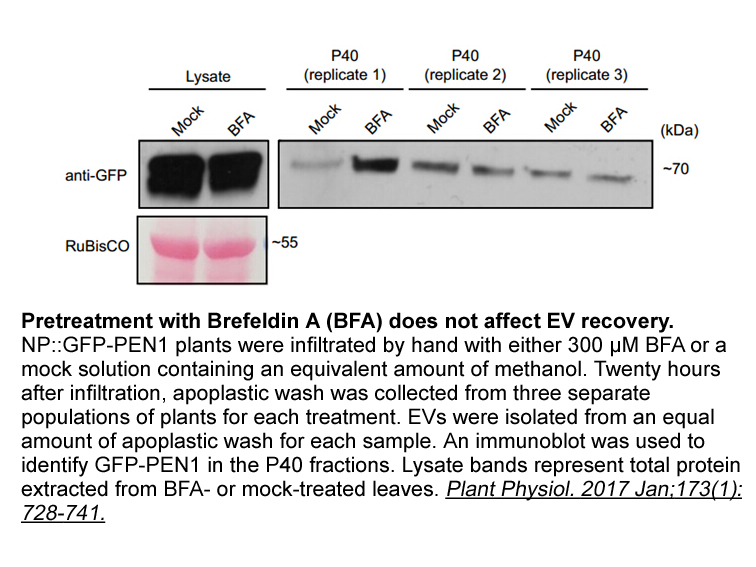
The Global Burden of Disease Study notes a substantial decrease in long-established global health threats such as communicable diseases and malnutrition, while underscoring the rise in non-communicable diseases and years lived with potentially disabling illnesses and injuries. One important but la
-
El ansia renovadora que suscita tambi n se crea y
2019-05-23
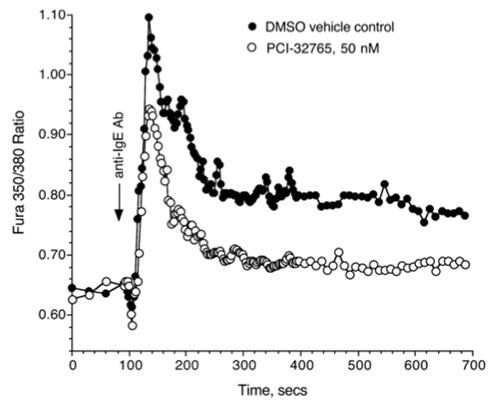
El ansia renovadora que suscita también se crea y re-crea en los paratextos ilustrados, que en cierto modo también sugieren una performatividad discursiva aplicada a la realidad convulsa de México en la revista Futuro, que puede observarse en la imagen y que se ha titulado, “Defensores del orden”.
16111 records 1031/1075 page Previous Next First page 上5页 10311032103310341035 下5页 Last page